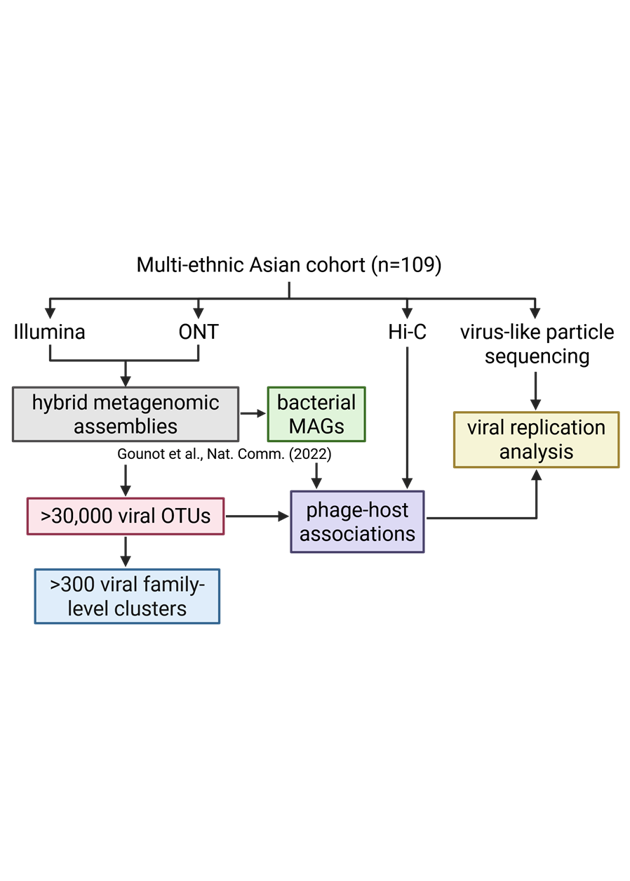
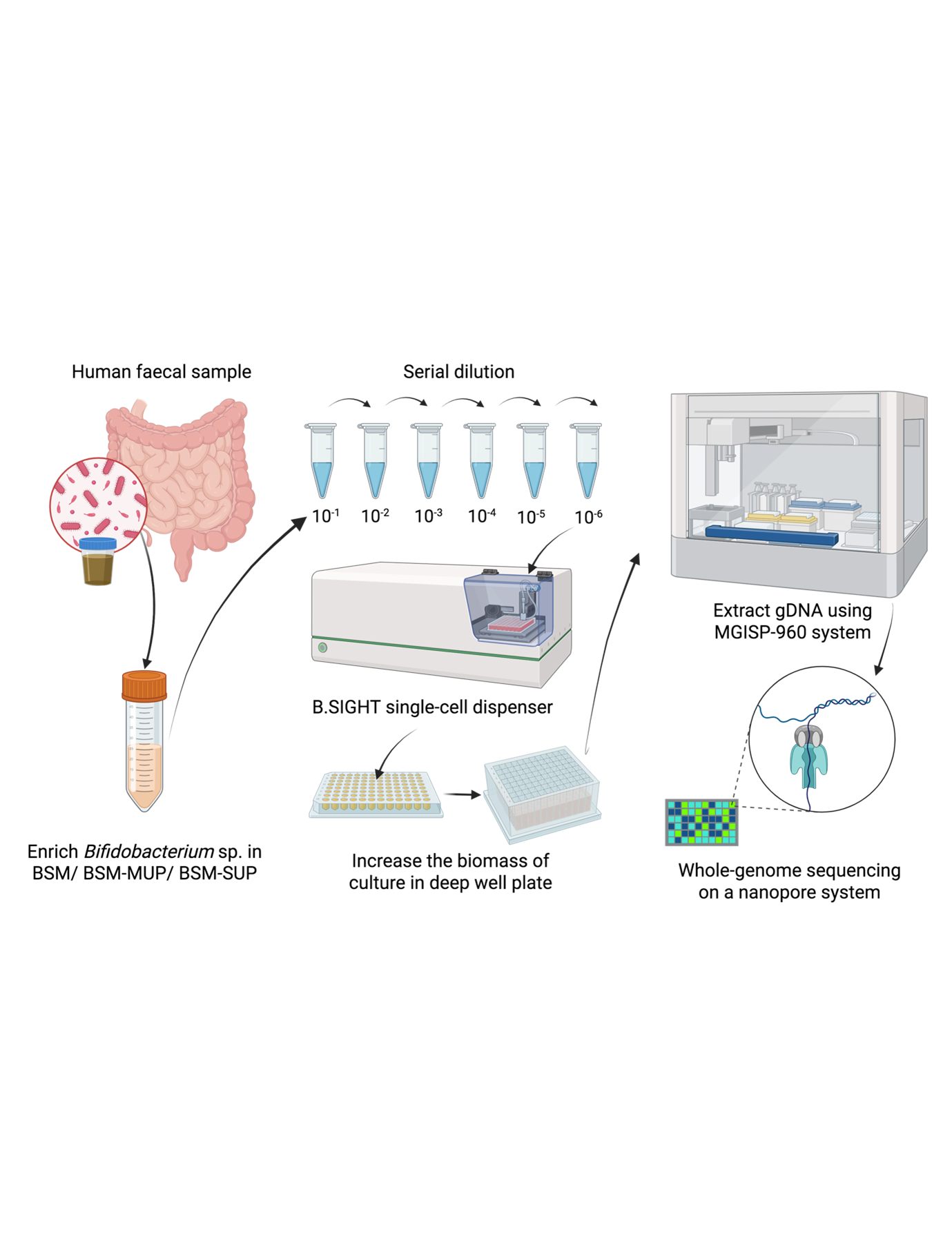
High-throughput microbial culturomics is an exciting field—but it’s still just getting started. One big challenge is: how do we efficiently isolate individual bacterial strains out of complex microbial communities?
By integrating a commercial single-cell dispenser (B.SIGHT) into a culturomics workflow, we made it much faster to isolate individual Bifidobacterium directly from faecal samples. Could we really recover isolates efficiently—and still capture substantial genetic diversity? It turns out that this is indeed feasible. Our work opens up new possibilities for understanding microbiome diversity, identifying probiotic candidates, and uncovering what Bifidobacterium function in the human gut.
So, how well does our workflow perform?
The B.SIGHT single-cell dispenser is highly precise, with only about 11% of microwells containing more than one cell, as measured using fluorescence signals. This means that most cultures are derived from a single parent cell as we would want.
Global diversity of a mock community was also preserved in post-dispensing cultures. When we dispensed a mixed community of nine species during their active growth phase, all species were still represented in the final culture, even though their relative proportions changed.
However, the best time to isolate bacteria depends on the species. We found that different Bifidobacterium species are most successfully cultured at different time points, highlighting the importance of sampling complex faecal communities at multiple timepoints to capture greater diversity.
To increase the likelihood of isolating the target species, we needed a highly selective medium for both enrichment and post-dispensing cultivation. Herein, BSM-MUP was found to be the most effective growth medium for isolating Bifidobacterium spp., outperforming other tested media and strongly limiting the growth of unwanted bacteria such as Enterococcus.
Using this approach, we readily generated over 600 bacterial cultures from faecal samples of five individuals in Singapore. These represent more than 20 distinct Bifidobacterium lineages—and over 60% are completely new compared to publicly available genomes!
Why does this matter? Because this scalable workflow does not have to be limited to Bifidobacterium. In principle, it can be applied to other microbial groups as well. By making it easier to recover microbes one strain at a time, this approach strengthens culturomics as a complement to DNA-based sequencing and brings us closer to studying what important microbes actually do—not just who’s there.