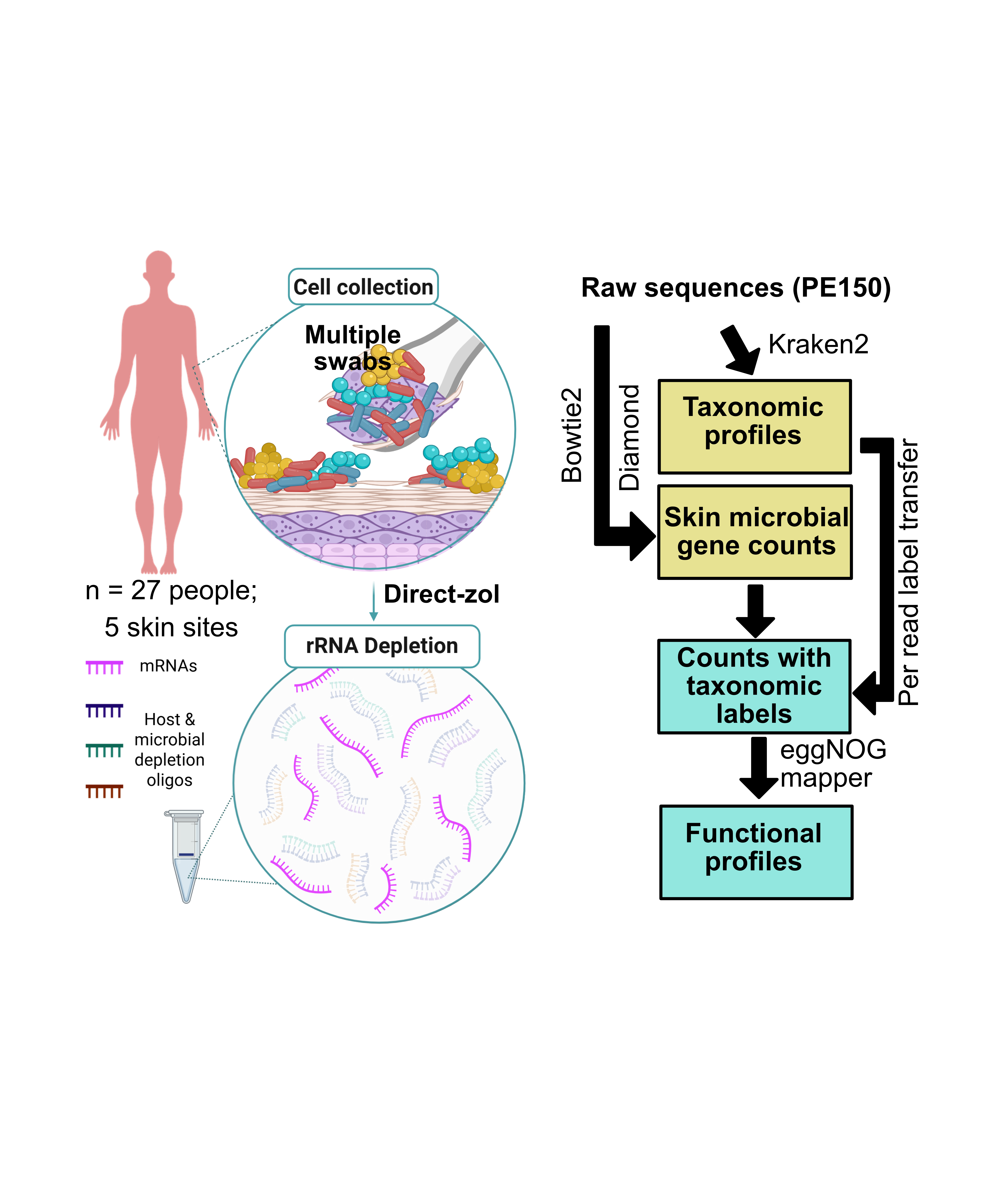
We are excited to share our latest work published in Nature Communications. In this work, we show how to achieve high-throughput sequencing of DNA containing non-canonical bases (NCBs) using nanopore sequencing and de novo basecalling enabled by splicing-based data-augmentation.
The inability to read non-canonical bases in a direct manner has been a significant limitation for applications in data storage, nucleic acid therapeutics, and synthetic biology. Here we demonstrate that such bases can be directly analyzed using a MinION sequencer to generate unique signals that can be deconvoluted by a specialized basecaller model.
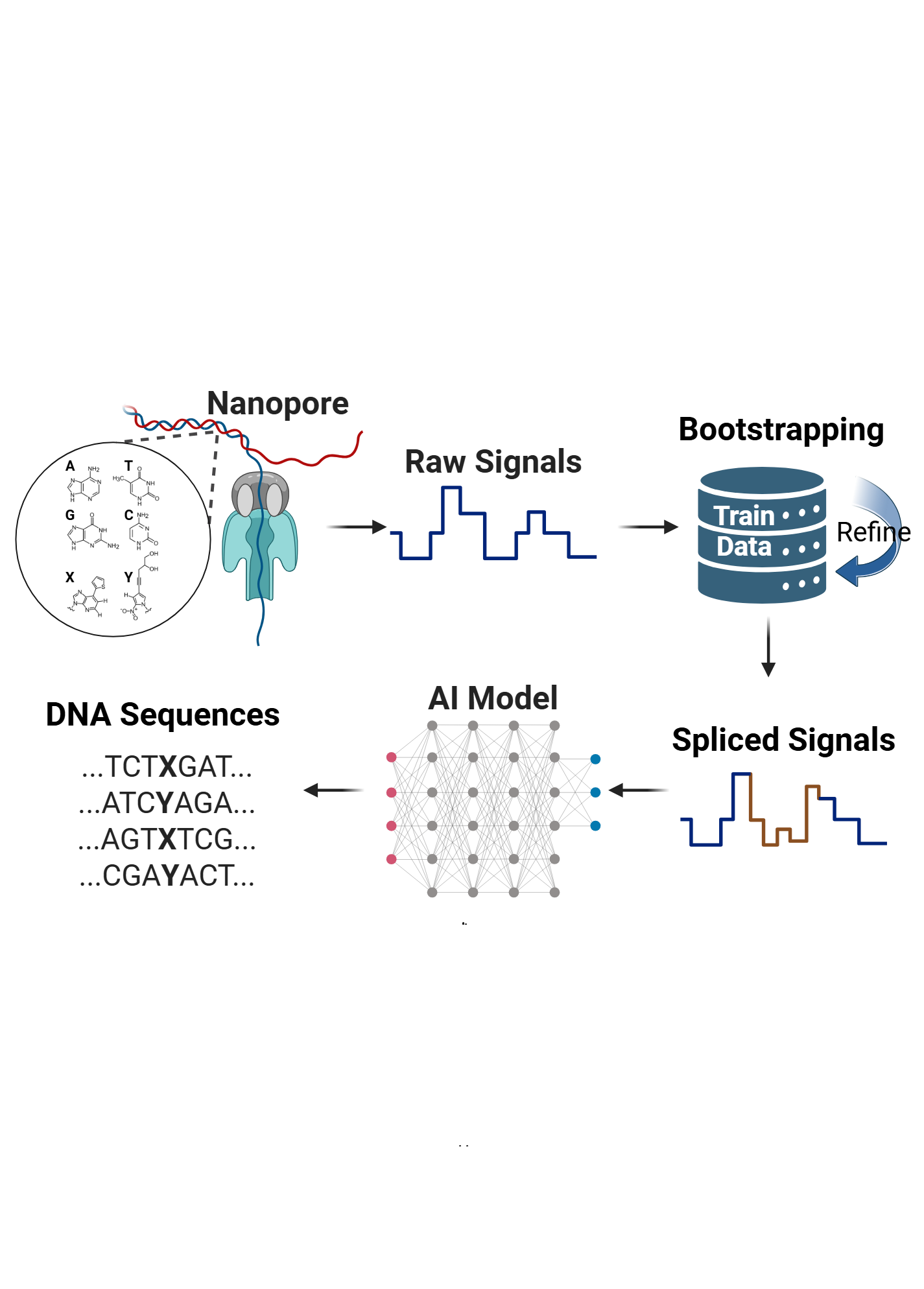
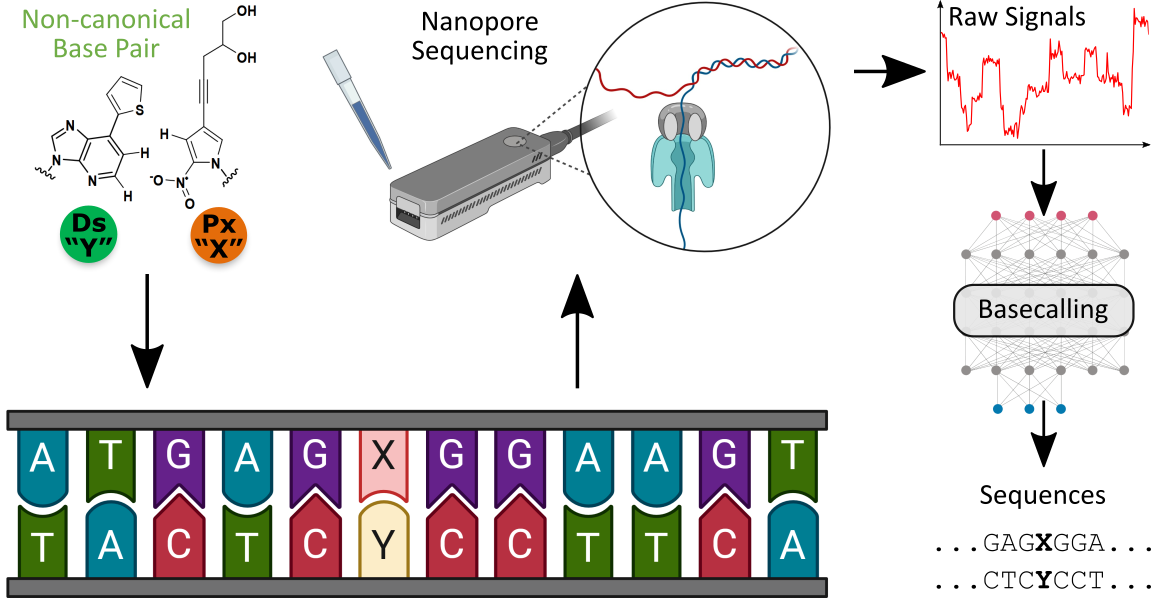 Figure: Overview of workflow, showing the synthesis of XNA templates containing non-canonical basepairs, nanopore sequencing, signal-level analysis and direct deconvolution of canonical and non-canonical bases.
Figure: Overview of workflow, showing the synthesis of XNA templates containing non-canonical basepairs, nanopore sequencing, signal-level analysis and direct deconvolution of canonical and non-canonical bases.
We validated that nanopore sequencing throughput is not impacted by NCBs and that there is notable distinguishability of their measured signals versus DNA controls and canonical bases.
To obtain a machine learning model that can deconvolve these signals and basecall NCBs along with natural bases, we developed an enzymatic framework to synthesize DNA with high NCB purity (>90%), and generate a library of 1,024 NCB-containing oligonucleotides representing them in diverse sequence contexts.
We leveraged our library by employing a data-augmentation technique based on splicing nanopore signals to generate reads containing NCBs in even more diverse contexts. Using these reads, we trained a generalizable model capable of sequencing NCBs with high accuracy (>80%) and specificity (99%).
Read all about this novel solution and results in our scientific paper and check our reproducible code on GitHub!