The human gut microbiome harbors an immense diversity of bacteriophages (phages) — viruses that infect bacteria — but their diversity and function remain under-characterized. For example, the most well-known gut phage families — such as crAssphage — infect the Bacteroidota phylum and are obligately lytic.
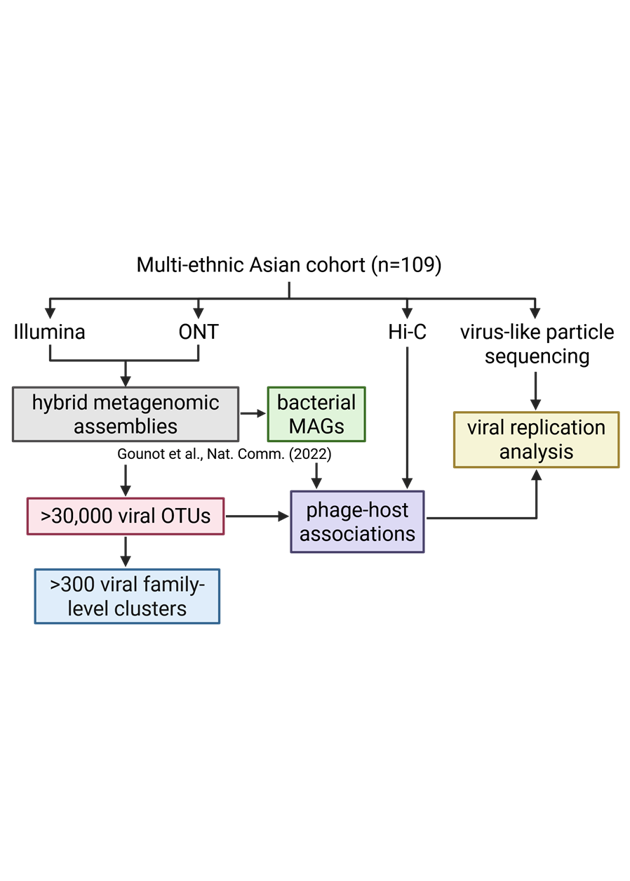
Leveraging long-read metagenomics, we mapped phage diversity in a Singaporean gut microbiome cohort (n=109), and performed proximity ligation (Hi-C) and viral-like particle (VLP) sequencing to paint a comprehensive picture of who they infect and their replication lifestyles.
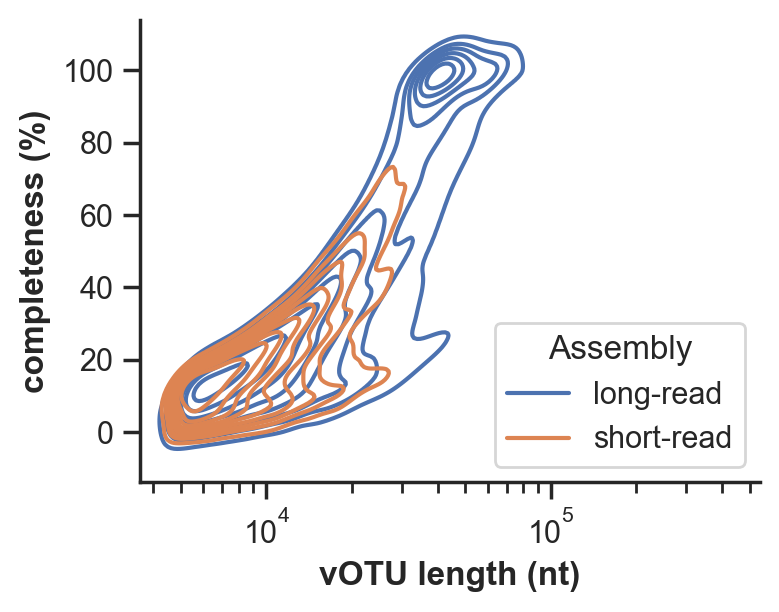
We found drastically improved viral recovery with the addition of long-read sequencing, and frequent fragmentation of viral genomes in short-read assemblies.
Viral family-level clustering revealed that almost all of the most prevalent gut phage families in this cohort have not been previously described in the literature (11 of the top 12).

A first guess was that these clades are population-specific and lurk only within Singaporean microbiomes. Surprisingly, we found that many of these families are in fact highly prevalent globally (>20% prevalence in >3,000 samples)!

Many of these families have broad Firmicutes host ranges; these so-called GuFi phages, which have so far evaded characterization, highlight how much there is yet to discover within our gut microbiomes.

Intriguingly, while many of these families are temperate — meaning they spend part of their life cycle in a dormant state integrated in the bacterial host genome — we find that some frequently induce and actively replicate. These results suggest underappreciated roles of these phage families in the gut.

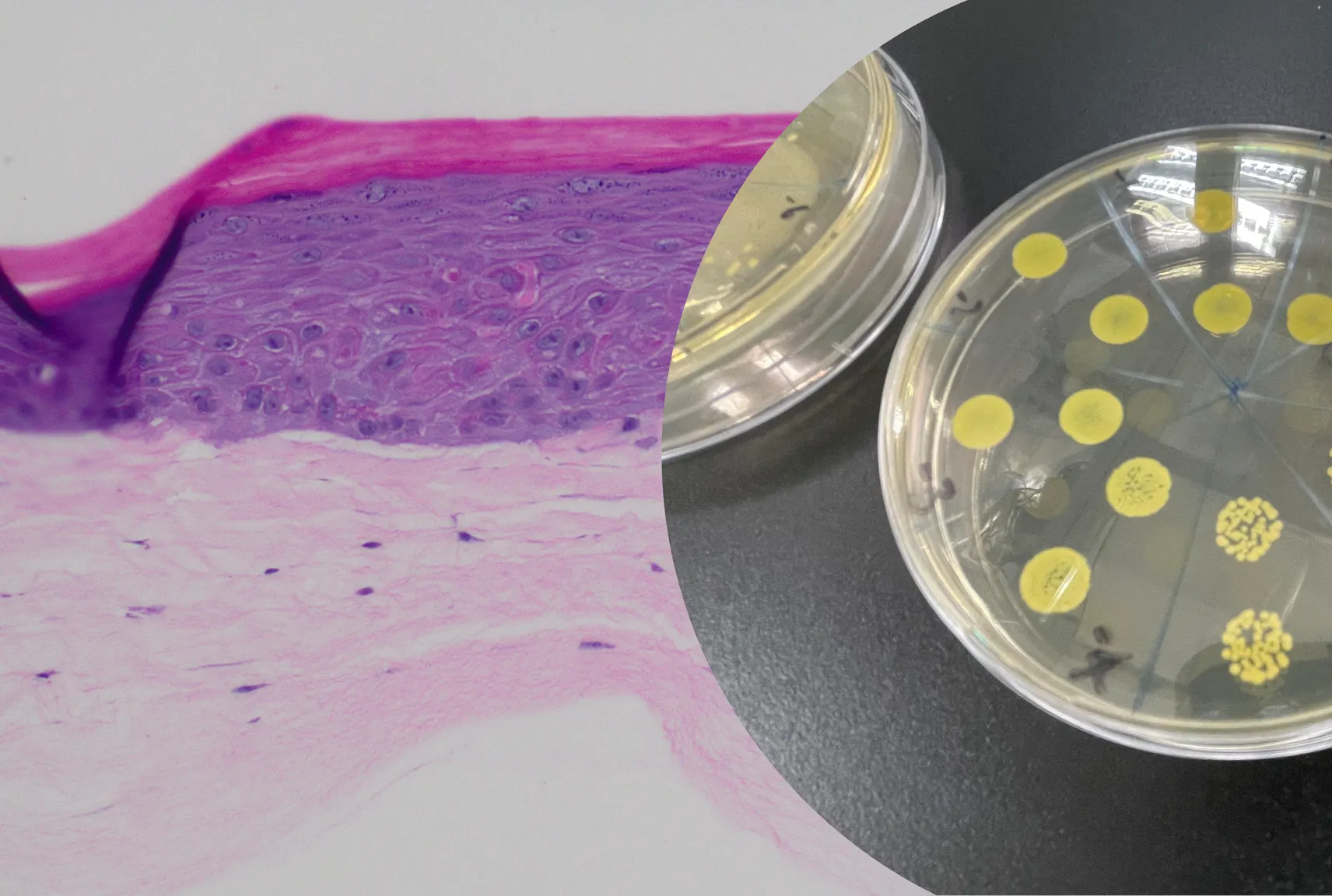
Our efforts have also led to the in vitro induction and TEM imaging of members of these families.

One big unanswered question is what explains the high prevalence of these phage families and not others? We explored many factors that could explain this, including the presence of diversity generating retroelements (DGRs), which aid in host range expansion and immune evasion.

Our study emphasizes the utility of long reads for viral discovery, and highlights globally prevalent phage clades that likely have a greater impact in the gut microbiome than previously appreciated.