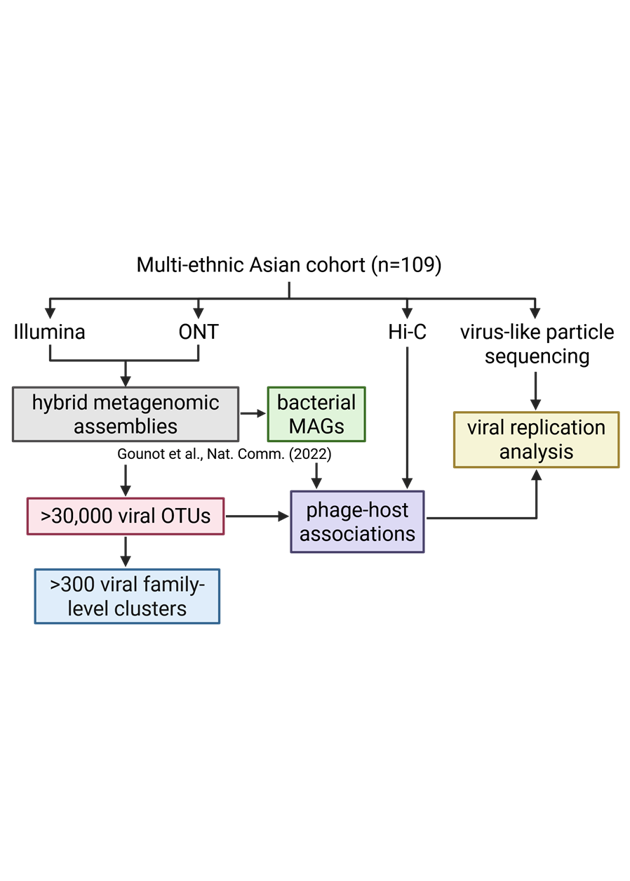
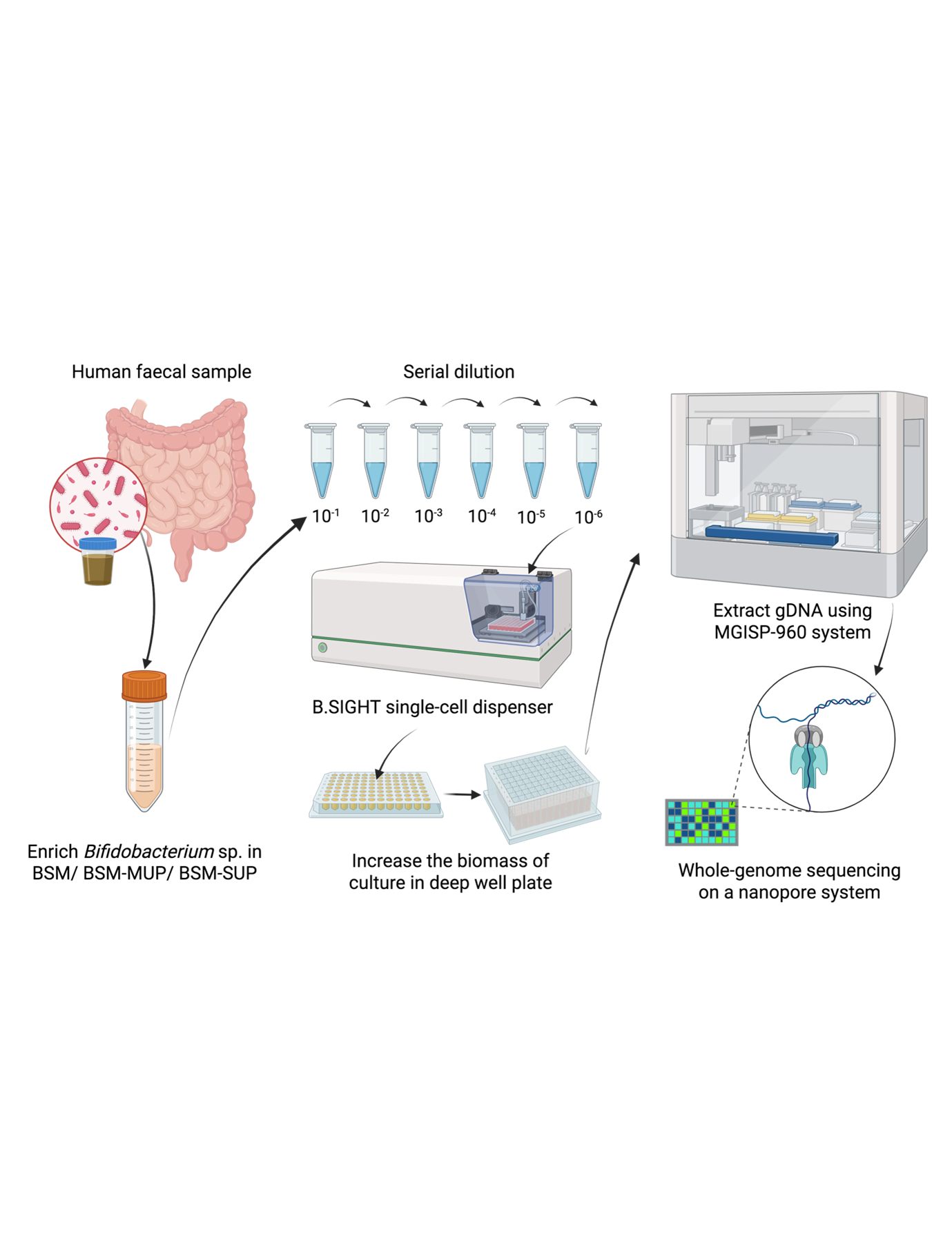
OPERA-MS: Hybrid Metagenomic Assembler
A hybrid metagenomic assembler which combines the advantages of short and long-read technologies to provide high quality assemblies, addressing issues of low contiguity for short-read only assemblies, and low base-pair quality for long-read only assemblies.